“Death and taxes” is a phrase that is usually attributed to Benjamin Franklin from a quote in a 1789 letter: “In this world nothing can be said to be certain, except death and taxes.” Public cloud computing providers didn’t exist back in the days of Franklin, but if they did, they would have no doubt made the list. Here’s why. Public clouds for large data analysis, just like death and taxes, are clearly inevitable because of two things. One simple and now rather worn out cliché. That would be scale and the slightly more subtle data.
Nation states are racing to provide their military and research organizations with access to exascale computing. Unfortunately, exascale is still benchmarked against outdated matrix diagonalization tests for tightly coupled computing performance. For example, the Linpack benchmark first appeared in 1979 as an appendix to the Linpack user’s manual. There is now a growing international obsession and competition to measure a system’s floating point computing power, judging and then ranking them by how fast a particular computer can solve a dense n by n system of linear equations. Billions of dollars are spent to solve Ax = b, and while all this is happening, the hyperscalers are winning.
The hyperscalers are winning because of the two reasons we stated earlier – scale and data. Decades of discovery have shown that more data produces better results. When you couple high quality, dense data with the right level of computational scale, you win. It is that simple. It no longer matters not one jot how fast you can solve for Ax = b.
The sheer amount of compute being added every hour to the exascale business outstrips any potential national of government level of investment. We live a long way from the 1960s post-war, government-level and Kennedy inspired moonshot era, and new economic dynamics are now at play. To quote Kennedy, we no longer “do the other things, not because they are easy, but because they are hard,” we have started to ignore many of the hard things, and most often even “the other things” as well. Mostly, well, because it is really hard, but also mostly because it just isn’t good business. For a capitalist society to function, you need capital.
Accordingly, and with ever-shrinking budgets, it is inordinately hard for academics to achieve any level of integrated compute at scale and then use it to solve difficult issues. The Next Platform looked into one of the most difficult issues in medical science – trying to work out why people die. Franklin said that death was inevitable, but we wanted to understand why we can’t predict it, and why academic leadership is being forced to turn to the hyperscalers for help.
Medical Research: Enter The Hyperscalers
In a recent paper in Nature titled, Scalable and accurate deep learning with electronic health records, the authors took Electronic Health Record (EHR) data from the University of California, San Francisco from 2012 to 2016 and the University of Chicago Medicine from 2009 to 2016. They selected out a total of 216,221 hospitalizations involving 114,003 unique patients. The percent of hospitalizations with in-hospital deaths was 2.3 percent (4930/216,221), unplanned 30-day readmissions was 12.9 percent (27,918/216,221), and long length of stay was (23.9 percent). Patients also had a range of one to 228 discharge diagnoses.
So, to summarize: there was an average of two hospitalizations per patient, 114,000 patients studied, 4,930 of them died in the hospital and for those fortunate to leave hospital they had between 1 and 228 potential diagnoses at their discharge. All of this is based on the medical records from two individual hospitals over a time period of between four and seven years.
We have spoken at length about unplanned data before. This paper is a canonical form of unplanned data, as the researchers stated “for predictions made at discharge, the information considered across both datasets included 46,864,534,945 tokens of EHR data.” To understand the impact of the required compute on this massive number of tokens that were found, the researchers added: “Moreover, our methods are computationally intensive and at present require specialized expertise to develop.”
Looking at the author list, not only Stanford University, UCSF and U Chicago research medical scientists are represented, but also specifically, 31 individual research and data scientists from Google’s Mountain View campus in California are listed. Yeah, 31 Googlers. That’s a lot of “specialized expertise” right there. Google has never been shy about its hiring practices and often speaks to bringing in only the best people it can into its teams. The Google Brain team is no different in this regard and is full of highly specialized talent.
In order to understand more about what the group stated as “computationally intensive methods,” we contacted the two lead authors of the paper to ask about speeds and feeds, and how the computation really worked behind the scenes. We gave them seven days to respond. We never heard back so we have literally no idea what “computationally intensive” actually means, other than there were 31 experts from Google to help publish this paper. Just in terms of FTE salary investment we have to imagine it was coupled with a significant amount of computational horse power for this research to play out. We recently dissected the third version of Google’s TPU TensorFlow platform, Google is not short on core computing capacity in anyway shape or form, but more on that later.
To dig in further, we examined the public code repository from the paper that explains how the FHIR format data was used and computed against. The FHIR format used in the paper is available at https://github.com/google/fhir. While trying to understand how the actual compute was assembled, we noticed, “The transformation of FHIR-formatted data to TensorFlow training examples and the models themselves depend on Google’s internal distributed computation platforms that cannot be reasonably shared.”
Also investigating the data in the GitHub repository we noted references to the Google implementation of protocol buffers for FHIR and the BigQuery platform. They do clearly state however that this is not an officially supported Google product. So Google’s servers and software were used extensively for this analysis, which is important to note as this study involves research and machine learning on sensitive EMR data from actual humans with varying levels of consent.
Healthcare Records
We examined the paper further to understand what was inside the healthcare records and found the following statement: “datasets contained patient demographics, provider orders, diagnoses, procedures, medications, laboratory values, vital signs, and flowsheet data, which represent all other structured data elements (e.g., nursing flowsheets), from all inpatient and outpatient encounters. The UCM dataset additionally contained de-identified, free-text medical notes.”
This is very detailed stuff, and clearly an incredibly rich and powerful dataset which was then tokenized in preparation for feeding into assorted neural networks. This tokenization generated 46,864,534,945 tokens for 114,003 individual humans or an average of 411,000 descriptive medical words per person, so approximately 100,000 words per person per year. To try to put these words in perspective, we downloaded one of our own Facebook profiles and counted up ten year’s worth of status update words that can be found in timeline.htm for that account. We found 70,000 tokens (7,000 a year) generated from that little box on Facebook that says: “What’s on your mind?” Medical records from this dataset are approximately 14 times more dense than what a slightly above average user posts as simple status updates on a Facebook page, not including comments on their friends posts, or instant messages, only “What’s on your mind” status messages. Reminder: Facebook installs an awful lot of servers to track the sentiment of their users.
So, 100,000 patients were indexed by top tier scientists and researchers on the Google cloud and then analyzed. The combined research team found to the best of their knowledge, that their models outperformed existing Electronic Health Record models in the medical literature for predicting mortality by 0.92 to 0.94 versus 0.91, and unexpected readmission by 0.75 to 0.76 versus 0.69. They continue to state that the models also outperformed traditional, clinically used predictive models in all cases.
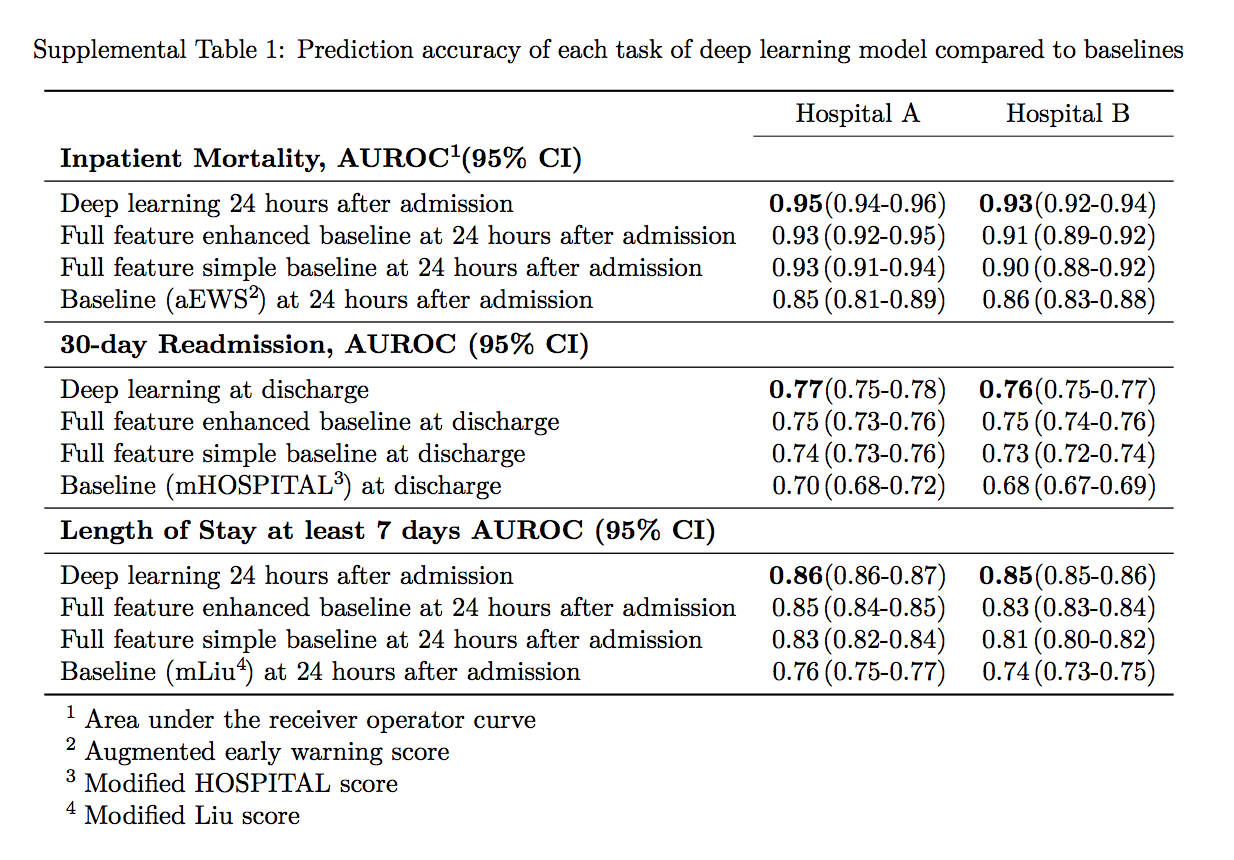 The study report continues with this statement: “We believe that this approach can be used to create accurate and scalable predictions for a variety of clinical scenarios. In a case study of a particular prediction, we demonstrate that neural networks can be used to identify relevant information from the patient’s chart.”
The study report continues with this statement: “We believe that this approach can be used to create accurate and scalable predictions for a variety of clinical scenarios. In a case study of a particular prediction, we demonstrate that neural networks can be used to identify relevant information from the patient’s chart.”
So in what is rapidly becoming a war of inches in the artificial intelligence community, 31 Googlers and unknown amounts of compute (but we know it to be significant) were able to bend the curve from 0.91 to 0.92 to 0.94 for predicting mortality, and from 0.69 to 0.75 to 0.76 for unexpected readmission. There is clearly an incredibly long way to go, but the more importantly, Google has started to gain legal access to the population’s medical data sets so it can continue to add medical data to its cohorts in order to further improve their methods. More data, better answers, more insight. There are walled gardens around all of this data. For now.
For decades, we have heard many research center directors and center managers – this author included – justify their roles and state that “sensitive data can’t possibly go on the cloud.” So how was this research completed? The paper is crystal clear and transparent about the whole issue of informed consent and data analysis: “Ethics review and institutional review boards approved the study with waiver of informed consent or exemption at each institution.”
To be more explicit, data transparency is achieved via collaboration with world class medical research scientists, but also more importantly through a “waiver of informed consent” via 45 CFR 46.116(d) and recent FDA Guidance:
- The research involves no more than minimal risk;
- The waiver or alteration will not adversely affect the rights and welfare of the subjects;
- The research could not practicably be carried out without the waiver or alteration; and
- Whenever appropriate, the subjects will be provided with additional pertinent information after participation.
Examples of each participating institute’s “informed consent waiver” can be clearly found at Stanford, UCSF and Chicago. Each IRB at each institute gave a waiver to the patient informed consent, and also allowed for storage of data and research output extramurally. Each of these top tier R1 research institutes have strong cyber infrastructure investments available physically on site, so the need to seek outside resources is even more fascinating.
This data movement and mobility fact alone could well herald in a new wave of advanced medical inquiry on consent based medical records that were traditionally off limits to so many researchers. It literally changes the shape of computing and data storage and network capabilities in meaningful and yet to be fully understood ways.
Importantly, these massive public healthcare computations may also end up being off limits for governments, and here’s why. Let’s assume based on this four year average study that there are around 411,000 tokens per patient, about 100,000 tokens per person per year. The 2016 census data shows 8.8 percent or 28.1 million people did not have medical insurance out of 320.3 million people (who filled in the census in 2016) in the USA. So, for the purpose of this study, we can safely assume the size of the problem is of the order 300 million healthcare records. This is 3,000 times the size of the nature study we have discussed.
This paper studied two specific issues, unexpected readmission and hospital mortality.
Using a 3,000 scale multiplier even at the most simple level for two predictable medical outcomes, this is clearly an exascale problem. Even with all the FTE resources, compute, and storage, the predicted mortality result was effectively 2 percent more accurate than the current state of the art to predict if you might die when you are in hospital. We don’t know what it costs to make this slightly more accurate prediction.
In the light of this early study, the question we must ask is, who will even have the resources, compute and available methods and technology to be able to accurately build and diagnose and future healthcare issues based on artificial intelligence?




Be the first to comment