
Building custom processors and systems to annotate chunks of DNA is not a new phenomenon but given the increasing complexity of genomics as well as explosion in demand, this trend is being revived.
Those that have been around in this area in the last couple of decades will recall that back in 2000, the then Celera Genomics acquired Paracel Genomics (an accelerator and software company) for $250 million who at the time had annual sales of $14.2 million. Paracel had a system they called GeneMatcher, who were able to fit 7,000 processors into a box that could compete with over 1,000 Pentium computers. It could (as many of these magic black box devices do) only compete against a few boutique applications, but those were the important ones, as they allowed for super fast hidden markov model sequence alignments that were required for genome annotation. It turned out these systems could also compare large amounts of text very quickly so there was an additional market in the government’s multiple three letter agencies for “text mining”, Again, very little of anything really changes.
So, it made sense at the time that sequencing commercial giant Celera (who were ultimately acquired by Quest Diagnostics in 2011) to pick up the accelerator company during the human genome race. Unfortunately, history hasn’t exactly been all that kind to the field of dedicated devices in genomics. Compugen for example dropped their accelerator products many years ago, although there are some holdouts. Timelogic for example, to this day continues in FPGA technology even after having been founded all the way back in 1981, and that’s an extremely long run for any company, especially in this space.
FPGAs have always been such an interesting and tantalising platform for sequence analysis due to their inherent and massive potential for parallelization. So much so, and in spite of all the history, new startups in the Bio / FPGA space such as Edico Genome recently sprang into existence. In 2014, Edico raised $10 million on a finance round led by Qualcomm Ventures and included Axon Ventures and Gregory T. Lucier, the former chairman and CEO of Life Technologies. Today, Edico marks another landmark by being acquired by the DNA sequencing powerhouse Illumina for an undisclosed sum. Clearly bringing back very strong memories to many in the bioinformatics community as they remember the Paracel / Celera days.
Edico had selected both Dell EMC and IBM as technology partners for their platform. Specifically Isilon, based on the large storage requirements of NGS and the obviously workhorse Dell R730 double socket servers, but also more interestingly the System S822LC from IBM’s Power division. A proverbial soup of technologies. Another area where Edico also made headlines recently was around their cloud based FPGA offering. As one of only a handful of publicly listed AWS FPGA customers, Edico were able to repackage their kit directly into the cloud and leverage the custom AWS F1 instances. Clearly Illumina are buying a whole bucket of technology parts and pieces here with this acquisition, it will be interesting to see how it plays out. “Undisclosed sum” acquisitions often have an aroma of the “equi hire” where companies are purchased for a combination of technology but mostly for their staff. It’s clear the number of people who can program FPGAs successfully are small, and Edico have proven that they can achieve it, which makes the staff inherently more valuable to the parent company.
Next Generation Sequence Storage
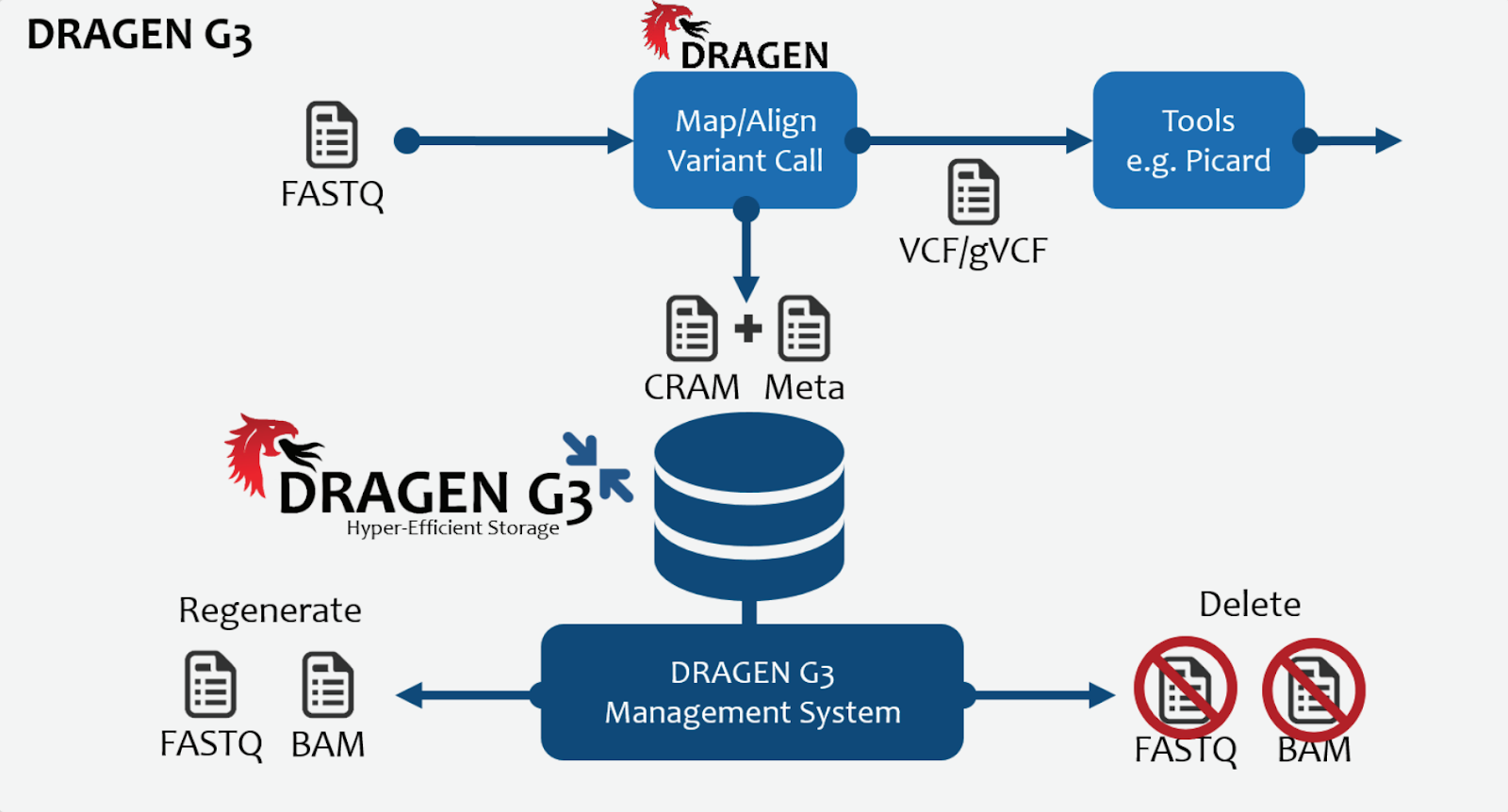
Interestingly, the DRAGEN G3 data compression software also comes along with this acquisition. Genomics has always been limited by two major issues. Large data sets, and large compute requirements, each non trivial. The FPGA toolkits clearly help with compute but G3 and data compression algorithms like it are of significant interest in NGS pipelines. With G3, Edico claim a 70% reduction in data size in lossless mode and 80% in quantized or lossy mode, also claiming to be able to crunch through a whole genome BAM file in about 8 minutes. This coupled with the chemistry and robotics expertise at Illumina will make for some interesting future products. How the interaction with partners at DNAnexus, and Seven Bridges will play out along with the AWS cloud story is going to be interesting to watch. BaseSpace Sequence Hub at Illumina now in theory, for example ought to be able to be quickly stitched together with the F1 and DRAGEN cloud offering as only but one simple example.
The community also appears to be excited by the acquisition, Anthony Philippakis, Chief Data Officer at the Broad Institute said in the Illumina release, “The scientific community should align around standards to maximize the impact of genomics in health. We are excited to collaborate with Illumina on approaches and pipelines for the analysis of NGS data. The Genome Analysis Toolkit (GATK) has been adopted by a diverse set of researchers, and we look forward to integrating these methods with Illumina sequencers to improve the overall efficiency of data analysis — enabling the community to more easily share and collaborate.”
Lather, rinse, repeat.
So is this just history repeating itself? Maybe not, as it turns out accelerator technology is still of considerable interest to many in lifesciences, and it’s not all about FPGA tech there’s a strong sprinkling of GPU in many systems and services today. As a counter example, The Next Platform spoke with Roger Pettett, Senior Director at Oxford Nanopore. Pettett recently presented at the NVIDIA GPGPU conference about Oxford Nanopore’s range of sequencing technologies, each are supported by ever more aggressive computing platforms.
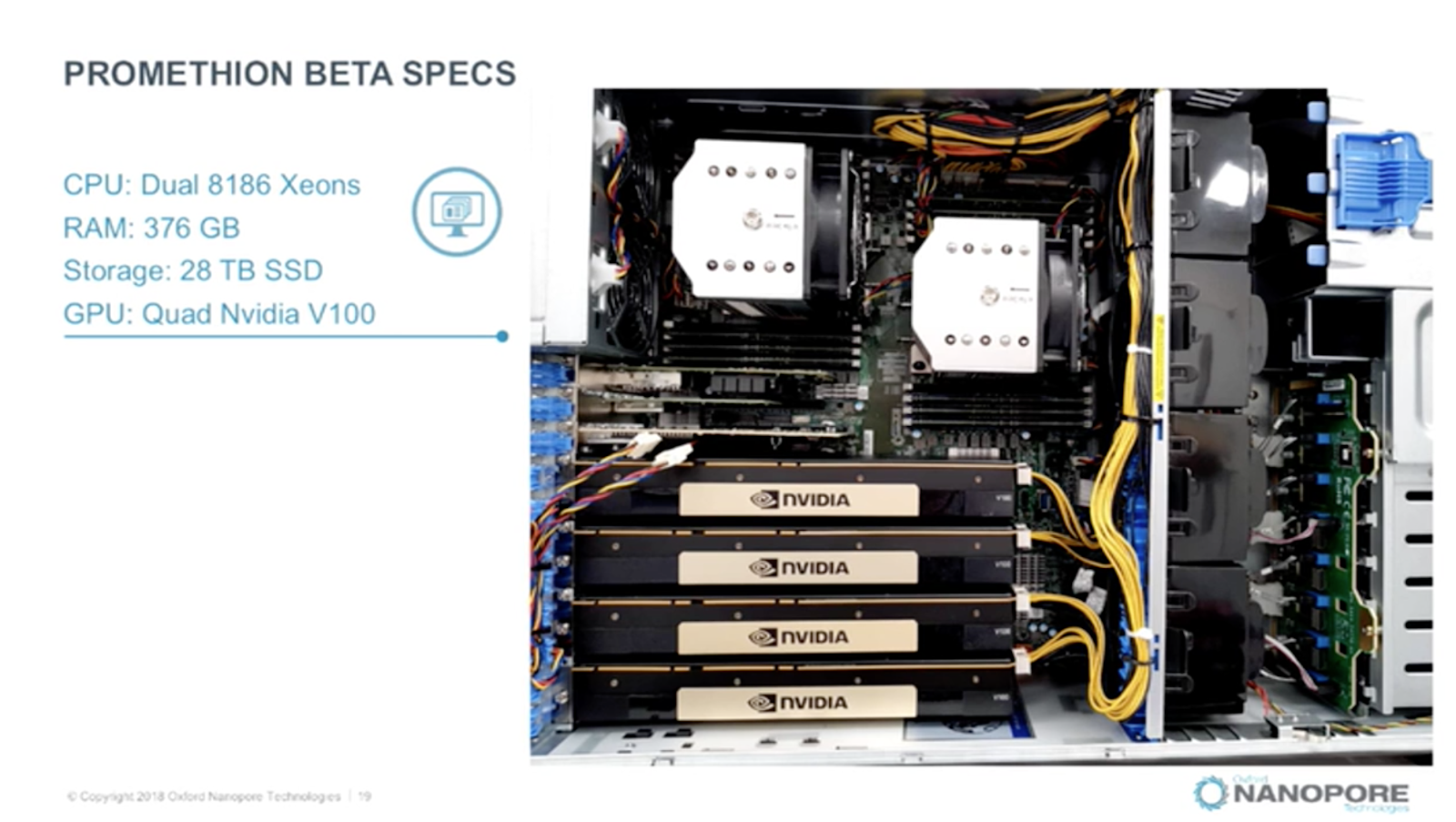
From embedded Jetson TX2 systems in the tiny MinIT product all the way up to the monster Promethion system that needs 4 Volta V100 GPGPU just to keep up with the sequencing rate. That’s an awful lot of compute to be sitting in or near the lab. However, from a distance this is also a dedicated system albeit constructed from more general purpose parts such as the V100 and Intel 8186 Xeons.
In summary, for additional background and context, The Next Platform spoke with Ewan Birney, Director at the European Bioinformatics Institute, who is also a compensated consultant to Oxford Nanopore. Birney summed up the state of the field nicely by saying, “Data intensive computational biology is starting to need dedicated hardware for a variety of applications.”
You can actually trace Birney’s dedicated device roots all the way back to implementing accelerated gene prediction and hidden markov models on those early hardware systems, now over 18 years ago. However, the point Birney makes here is key, and albeit that history has been somewhat unkind to dedicated devices, there is still a huge amount of interest in advanced, massively parallel accelerator technology buried deep inside sequencing and analysis pipelines and systems but also at an increasing number of companies, academic centers and research and development labs. New dedicated hardware histories are being made.







Be the first to comment