
Novel architectures are born out of necessity and for some applications, including molecular dynamics, there have been endless attempts to push parallel performance.
In this area, there are already numerous approaches to acceleration. At the highest end is the custom ASIC-driven Anton machine from D.E. Shaw, which is the fastest system, but certainly not the cheapest. On the more accessible accelerators side are Tesla GPUs for accelerating highly parallel parts of the workload—and increasingly, FPGAs are being considered for boosting the performance of major molecular dynamics applications, most notably GROMACS as well as general purpose, high-end CPUs (Knights Landing and other manycore architectures).
According to the leads behind a new effort toward a MapReduce accelerator with FPGA acceleration, even with the current general purpose accelerators, only part of the workload gets a speedup. Around 75% of an application like GROMACS can benefit from acceleration but that 25% can be targeted with a parallel accelerator like the one they have built and demonstrated with around a 4X improvement (on that non-accelerated section of the work).
 The team found that the degree of parallelism achieved by their accelerator for GROMACS was 60% for the compute-intensive force computation and 80% for the also intensive neighbor search with an overall 64% parallelism for periodic cell structure. To put this in perspective, existing accelerators focus only on the force computations, leaving performance on the table.
The team found that the degree of parallelism achieved by their accelerator for GROMACS was 60% for the compute-intensive force computation and 80% for the also intensive neighbor search with an overall 64% parallelism for periodic cell structure. To put this in perspective, existing accelerators focus only on the force computations, leaving performance on the table.
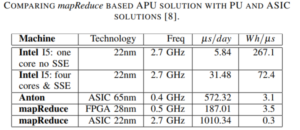
The developers say that the comparison with the Anton architecture from D.E. Shaw implemented as ASIC is not far off. “Our FPGA implemented programmable approach has performance in the same range with this specialized circuit, which is only 3X more efficient” although they add that the core technologies are not the same.
“A good compromise performance vs. price is provided by using hybrid computation solutions embodied in Accelerated Processing Units (APU). An APU integrate in the same system a PU with an accelerating parallel engine, such as a GPU, a MIC or, increasingly more frequently, a FPGA. The small accelerations provided by the use of the current parallel accelerators are mainly due to the fact that only the last two of the previously listed steps are submitted to the parallel accelerator, while the first step – the neighborhood search – which is theoretically the most time consuming, is left to the PU. Usually, the weight of the first step is minimized by the compromising solution of applying it only once at 10-50 state up-dates.”
Their approach also has some impressive energy efficiency figures, with 20X for the FPGA version of the MapReduce accelerator and 300X for the ASIC version. They note that there is a promising compromise between FPGAs and ASICs in implementing MapReduce accelertors for GROMACs in the eASIC technology, which provides a faster, cheaper way to bring new devices to bear.
The current version implemented on FPGA will give way to an eventual evaluation of 32nm technology. “The FPGA technology evolved from a pure programmable structure to a mixed solution putting together frequently used ASIC blocks with the in-field programmable blocks used mainly for interconnections. Besides the usual configurable logic blocks, the designers have access to memory blocks, DSP units, advanced microcontrollers (for example the dual-ARM Cortex 9) and a lot of standard interfaces.” The team says that “such FPGAs could be a wonderful support for a hybrid computing system centered on the microcontroller and based on the MapReduce accelerator developed on the programmable part.”
Even for those that have problems that fall far outside the molecular dynamics range, watching what happens here architectural is interesting because many of the same pressures are present in other applications and subsequent ASICs. In short, general purpose accelerators have demonstrated parallel pushes but these don’t extend throughout the entire application. With cheaper and faster access to ASICs we can expect a broader push that moves ideas like this one outside of the research lab and into potential production.


Be the first to comment