
Last week we described the recent, explosive growth of Amazon Web Services, which the company says is due in large part to an influx of new users—and existing AWS customers who are scaling their workloads ever higher. Although there are several markets driving this uptick in cloud usage at Amazon, the life sciences segment has been a target for AWS since the beginning and the company is seeing some interesting use cases and research efforts to keep pushing it further along.
Over the last few years, AWS has highlighted a number of large-scale genomics studies on their infrastructure, including the 1000 Genomes project and more focused genomics research efforts from companies like Novartis.. To attract more, they have beefed up their regulatory appeal to make sharing personal health data (not to mention securing it) less of a concern for this privacy-aware market.
While these cases showcase the potential for genomics on AWS, researchers in the area have managed to get the data collection step in the DNA analysis pipeline down to a science. In other words, getting the data to the cloud is not necessarily a challenges. However, when it comes to efficiently processing the petabytes of sequence information to make meaningful connections in a way that demonstrates the often-touted economic and time to result benefits, there are still some shortcomings, at least according to researchers from Harvard Medical School, Stanford Medical School, and a number of other institutions.
For most large-scale genomics research, the problems can be parallelized nicely across as many compute engines as teams can throw at the application, but optimizing at extreme scale and developing a workflow are still challenges. To this end, the team of researchers tapped the Amazon cloud for a scalable framework to test the limits of genomics workflows. Supported by a grant from Amazon Web Services, they were able to show how a custom workflow they developed was able to reduce the cost of analysts of whole-genome data by 10x.
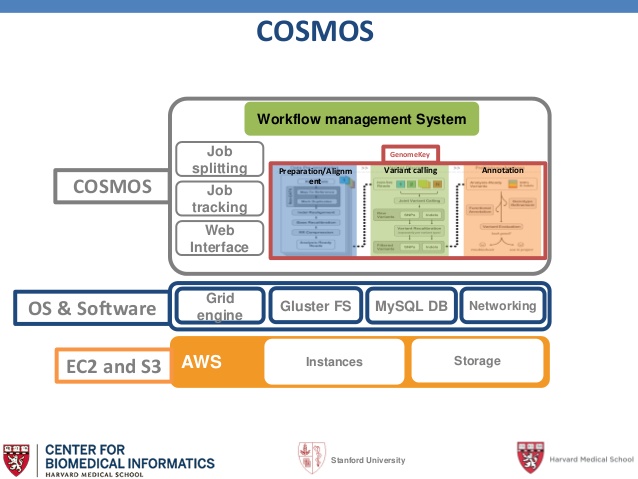
At its core, the project, called, COSMOS is a Python library designed to handle large-scale workflows that require a more formal description of pipelines and partitioning of jobs. Among the components are a UI for tracking the progress of jobs, abstraction of the job queuing system (to allow interface to multiple queuing systems) and fine-grained control over the workflow. While it has been benchmarked most extensively (and publicly) on AWS, it also runs on Google Compute Engine and on on-premises hardware.
COSMOS, tackles the problems that plague genomics workflows in two ways. As the researchers note, this happens via the implementation of “a highly parallelizable workflow that can be run quickly and efficiently on a large compute cluster—and can take advantage of AWS spot-instance pricing to reduce the cost per hour.” Target medical breakthroughs include genetic insight into the exomes of epilepsy and myelomonocytic leukemia in addition to other broader genomic studies.
The performance tests were done using standard AWS EC2 nodes in conjunction with the MIT-developed StarCluster manager, which handles provisioning and closing down of nodes based on the job requirements. Also at the core of the approach is Grid Engine, which is managed through StarCluster and is present on each of the AWS nodes. The COSMOS framework also has a plugin that allows installation of the GlusterFS file system on each of the nodes.
The work has been going on since 2013, but is continuing to showcase the potential for clouds in genomics. COSMOS was developed specifically to support the clinical-time, next-generation sequencing (NGS) variant calling pipeline, but can be used to develop pipelines for any large-scale scientific workflow. It runs the parallelized workflow for optimum and scalable processing of NGS data (currently whole genome and whole exome) and supports projects such as the 6000 exome and genome analysis.



Be the first to comment