![ruby-sat-6A2A8616[1]](https://www.nextplatform.com/wp-content/uploads/2020/11/ruby-sat-6A2A86161-1030x438.png)
The HPC world, particularly in the U.S. is waiting for the next series of transitions to far larger machines with exascale capabilities. By this time next year, the bi-annual ranking of the top 500 most powerful systems will be refreshed at the top as Frontier, El Capitan, Aurora and other DoE supers come online.
In the meantime, there have been several pre-exascale systems that appeared more recently across the U.S. national labs, not to mention several research workhorses that sported true architectural diversity, from Arm-based processors, to the latest GPUs from both Nvidia and AMD, not mention AMD as a rising CPU force in HPC or the new wave of AI-specific accelerators.
Lawrence Livermore National Lab has been at the forefront of all of those architectures over the last year in particular across systems like the just-announced Mammoth machine, the expanded Corona supercomputer, the existing Sierra and Lassen supercomputers that are paving the way for El Capitan, and as of today, yet another machine in the lineup, Ruby, a top 100-class all CPU (Intel Platinum) super, which we’ll get to in a moment. This is all coupled with recent investments in AI accelerators, including a SambaNova rack attached to Corona and the wafer-scale Cerebras machine attached to Lassen.
All of these developments made the lab one to watch this year from an applications point of view, but it was the addition of so much new hardware that kept them at the front of the news cycle throughout most of the year.
While the center was already planning around AI acceleration for its cognitive simulation aims and had a number of clusters in the works, it was the coronavirus funding that really stepped up their HPC ambitions across a number of machines. The CARES Act allowed for expanding the size of Mammoth, the Corona supercomputer’s expansion (the system’s name is coincidence, by the way), and the same goes for the newest addition, Ruby.
“We took a different approach than the other three labs that got CARES funding. Some brought in a single system to hit a small portion of their COVID workflow. We went out and talked to application teams to see what they were missing and brought in systems and balanced them to address different pieces of the overall workflow for coronavirus instead of just dropping a single system on the floor,” Trent D’Hooge, HPC integration and systems administrator at LLNL, tells us.
By being clever about creating a cross-machine workflow for their part of the CARES-funded research, the team was able to get maximum bang for buck out of several machines, adding to some while also using the opportunity to do interesting things for some backend operations, including work on their file systems and data management platforms, as described in a bit more detail in our description of the recent Mammoth machine. Mammoth was part of the $8.7 million in CARES Act funding that Lawrence Livermore received to help fight the coronavirus pandemic earlier this year. These funds were used to build out the “Corona” supercomputer, which we wrote about here last month as well.
“We were strategic about the funding and could partner with AMD and Intel to make our dollars go farther than might have happened with others. In the background we had our cognitive simulation and the normal big ticket things like El Capitan percolating in the background,” Ian Karlin, a computer scientist involved with several of the new machines, adds.
The uniqueness of LLNL’s distribution of the CARES funds allowed the lab to keep at the forefront of HPC on multiple fronts, enabling upgrades and further allowed them to fine-tune key parts of the CARES-related Covid research workflow to the right architecture given the range of options now available across AMD, Nvidia, Intel and other AI specific accelerators.
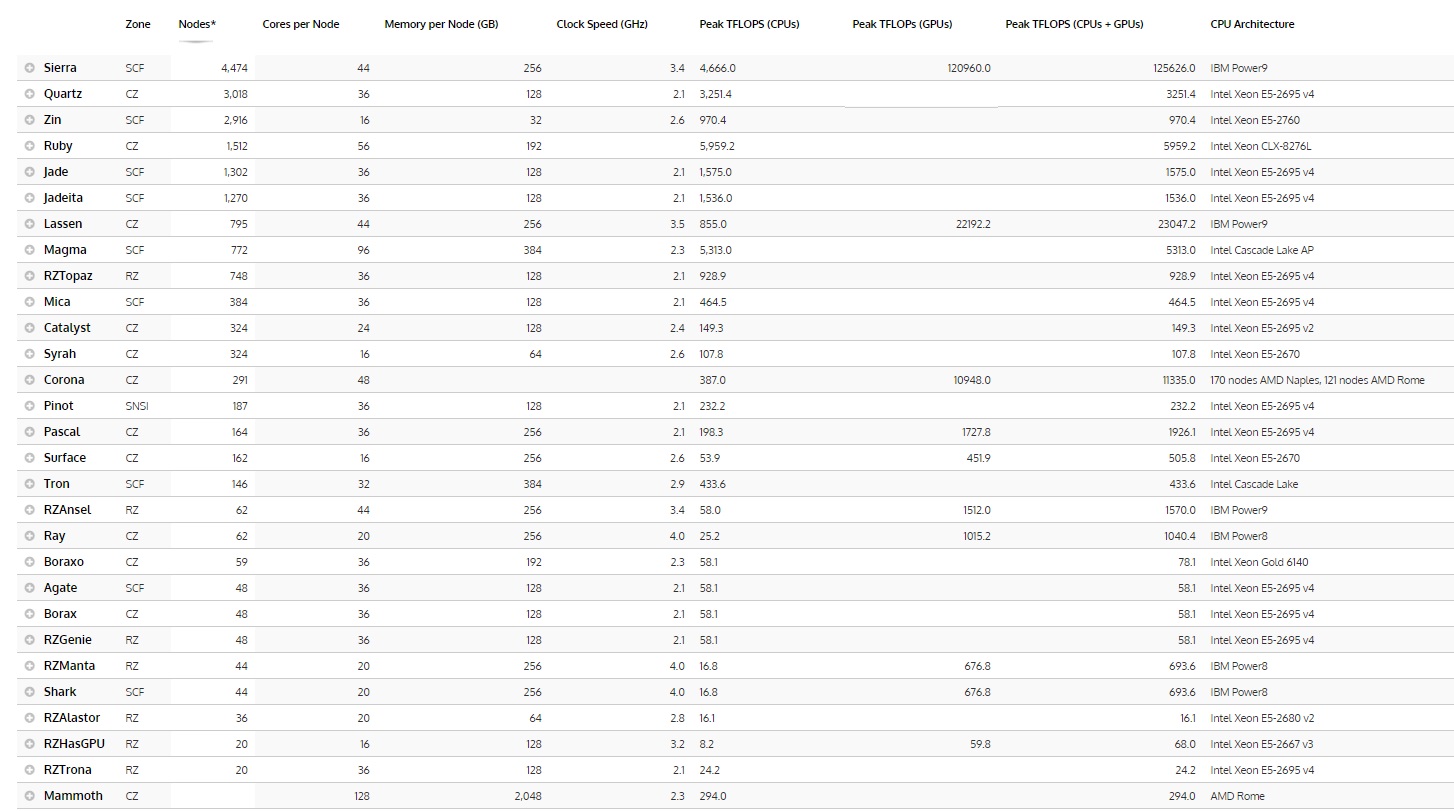
Below is an overview of the computing resources at LLNL overall, sorted by the most powerful in terms of capability, with Mammoth at the bottom due to a lack of updated information for this chart. Click to open in full.
The new Ruby cluster, pictured at the top, is a sizable workhorse. D’Hooge says they had it up and running LINPACK in time for the Top 500 announcement in a few days within a single week, which should place them in the top 100. There’s nothing fancy about the system, necessarily, other than the Asetek water cooling, which involved adding in a secondary loop via a Motivair CDU with 1.5 megawatts of cooling capacity) with Supermicro and Asetek figuring out the optimal cooling configuration. The datacenter where Ruby has support for 80kW per rack, but so far teams are only using 60.
LLNL COVID-19 researchers also have begun using Ruby to compute the molecular docking calculations needed for discovering small molecules capable of binding to protein sites in the structure of SARS-CoV-2 for drug discovery purposes.
“Ruby is excellent for running the molecular docking calculations,” said LLNL Biochemical and Biophysical Systems Group Leader Felice Lightstone, who heads the COVID-19 small molecule work. “Our early access on Ruby is allowing us to screen about 130 million compounds per day when using the entire machine. As our COVID-19 therapeutic effort moves toward optimizing compounds we have identified as promising, Ruby allows us to maximize the throughput of our new designs,” said Chris Clouse, acting program director for LLNL’s ASC program.





Be the first to comment