
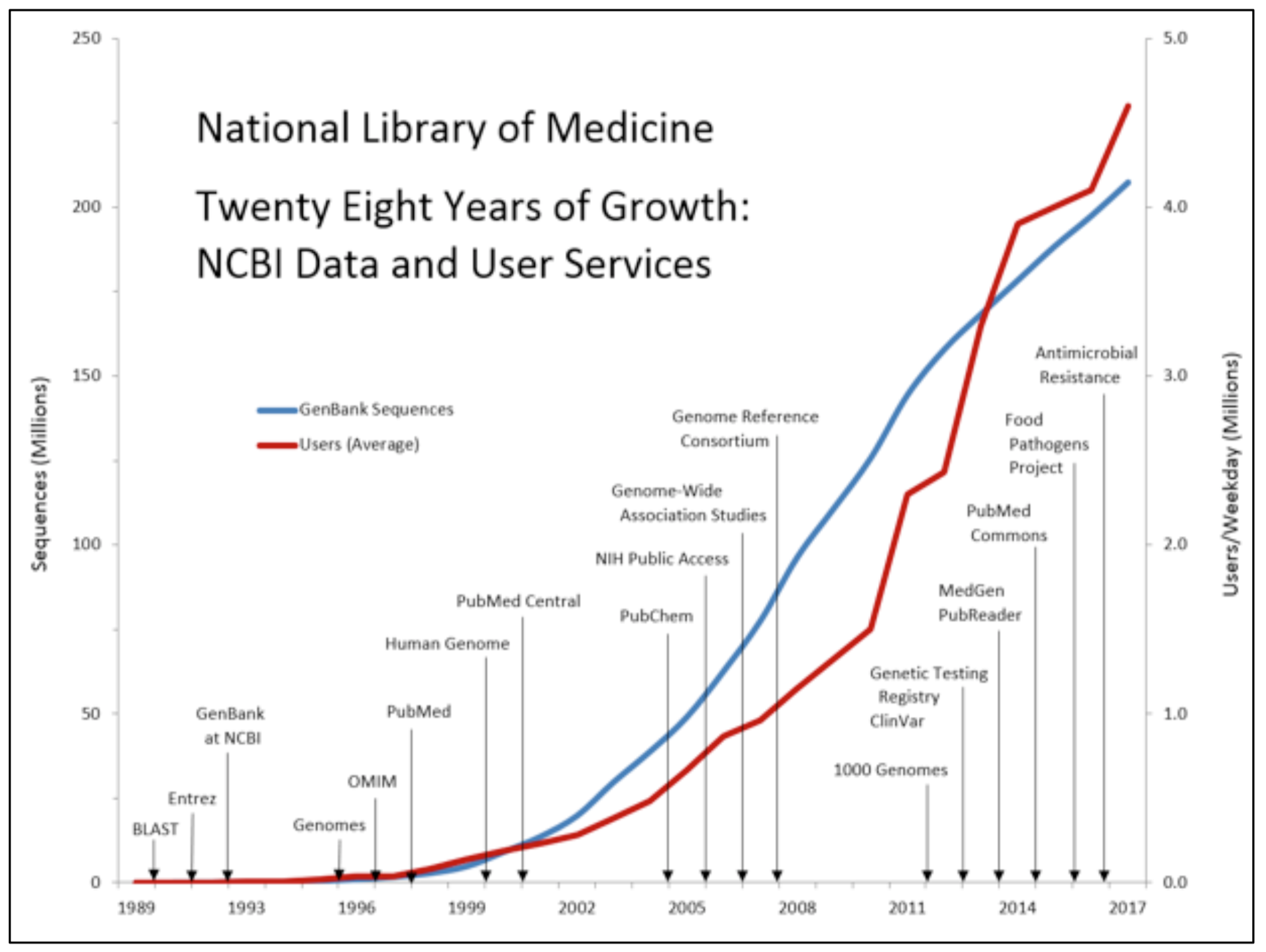
There is no shortage of data in the life sciences. Every talk about bioinformatics and genomics has to include the now ubiquitous hockey stick growth graph of digital DNA. It just isn’t a real-life science talk if there isn’t one of these graphs. These graphs have basically become analogous to singing the national anthem at the start of a live sports event.
As an example, The Next Platform recently attended the Broad Institute’s software engineering retreat to talk about our favorite topic of practical computational balance, joining Anthony Philippakis, chief data officer at the Broad Institute who kicked off the gathering with his own rather rousing anthem of terrifying hockey stick growth graphs. To fully highlight the issue, Philippakis continued to explain that in the last decade we had essentially “only just been collecting data.” The gathered audience of software engineers gasped, here we go, they thought – so what is he going to say next then?
“The next ten years” Philippakis explained, “will be critical because data will not only continue to be collected at an ever-faster rate, but we will also need to compute against all of it. At the same time.” This is the computational equivalent of walking while chewing gum. It wasn’t hard to imagine Bachman-Turner Overdrive belting out “You Ain’t Seen Nothing Yet” in the background as Philippakis continued to explain his vision of this new world of even further life sciences data complexity to the assembled tech crowd.
The team of software engineers had been brought together to share their own work, but also listen to senior experts like Philippakis and folks such as Andy Palmer, who is a seasoned entrepreneur, recently exiting Vertica to Hewlett Packard Enterprise. Palmer is a global expert in massive high speed data management at scale, most recently putting his money where his mouth is and founding the high technology data science company Tamr. Palmer spoke in detail to the engaged technology crowd about the challenges of his theories on DataOps and the challenge of data engineering at real world scale using decades of experiences in the massive pharmaceutical industry to further help annotate his story.
Afshin Beheshti, who is a bioinformatician and data analyst at NASA also presented and spoke about advanced genomics and the DNA analysis of space mice. Mice, literally in space. Testing out the effects of weightlessness on their genomic material while also being simultaneously bombarded with cosmic radiation. It is critically important research if we are to ever send humans further into space. Interestingly, the mice it turns out, adapt fairly quickly to their new environment – however, floating about with no gravity must initially be somewhat surprising and confusing to your average lab mouse.
For all of the gathered talent, (and space mice) there was something significantly more important going on during that retreat of highly skilled software engineers. Philippakis is the Broad’s chief data officer, but is also an MD and a PhD and is not a software engineer by training – that point is important as he hit the core issue head on during the introduction. Philippakis became superbly focused and carefully explained to the room full of software engineers that if the Broad is to be successful in the coming decades, software engineering must be considered as a first class citizen in the scientific process. It was a bold statement, you could almost feel that everyone in the room immediately considered themselves significantly more valued the very instant those words were uttered.
First Class Citizens Of Science
We could not agree more about how important this statement is. Today, without a well executed software and data strategy, essentially the entire modern scientific method just simply falls apart. It is also critically important that the MD and PhD voices in senior C-level leadership positions are talking directly to their staff about this issue. Gone are the days of the mostly ignored “expert nerds in the basement” approach to hiring and supporting bioinformatics and data science groups. Even during the heady heights of the work to inform the human genome, many software, computational and informatics groups were always considered to be somewhat lower down in the scientific social pecking order. Those folks, they do weird things with computers, they aren’t really quite actually scientists are they? Fortunately, that old, outdated pseudo-hierarchy is slowly changing shape – and across all areas of science. Computing is important, and how we physically compute will become even more important.
The informatics and software teams are finally being let out of the basements and the far flung portacabins, and welcomed into the esteemed laboratories as ever more importance is placed on their skills and vital in-silico methods. Bona fide professions are now finally being built around data science and research software engineering. The Broad Institute has become a leader in life sciences research and they also are directly following suit of this important trend by building out a new profession of software engineering to sit directly alongside and partner more fully with the “real scientists.”
Raymond Coderre is the director of people development and operations at the Broad. Coderre partnered up with Jeff Gentry who is an engineering manager for a number of high quality software teams at the Broad to work together to put on the event. This coupling of roles is the exactly the right approach to both increase the visibility and the importance of software engineering in life sciences.
We will need significantly more of this type of vision in many more institutes across the globe as exascale systems become more readily available, and yet increasingly complex. Here at The Next Platform we have always discussed the critical importance of not only single pieces of advanced silicon, but also the need to consider a stack or the whole platform. Humans sit on the top of that stack of parts, and each of the complex set of pieces needs to be orchestrated with absolute care, it’s not just about the tin and silicon. The humans matter.
Data Science Strategy At The NIH
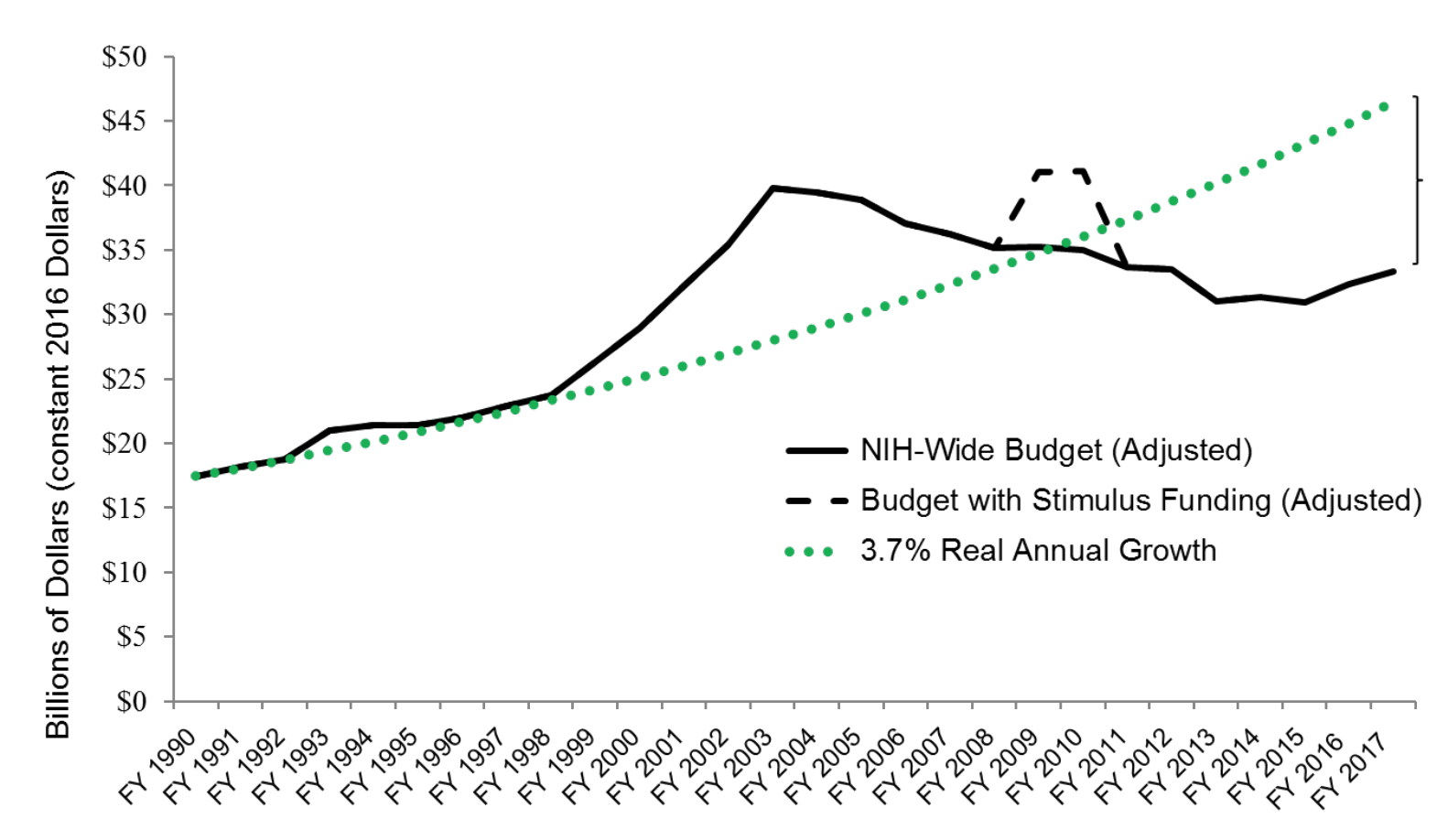
It’s not just the Broad. The world’s largest biomedical research agency, The National Institutes of Health (NIH) are also planning a new path forward. Recently releasing its Strategic Plan for Data Science, the NIH sets up a large number of stakes in the ground carefully marking out the area to pour a solid foundation for their data science activities. The NIH really does have to get smart about this for one major reason. There’s significantly less and less cash being injected into their system each year. See how this graph of budget below doesn’t match those hockey stick trajectories once the excitement of the initial human genome had died down? That’s a problem.
The NIH is not a single institute, there are 27 different centers and institutes that make up the whole, the organizational complexity is staggering, as is the amount of computing that needs to go on. Bioinformatics and genomics underpins such a large number of activities in life sciences that not having a well defined data science plan would be absolutely disastrous for the $37 billion giant organization. To put this in perspective, the NIH as a whole effectively every year spends the total assets under management that make up the Harvard University endowment. That’s a lot of money, and a lot of complexity and people. Unfortunately to reduce costs, leadership is also suggesting reducing the maximum amount of a salary payable with NIH grant funds from $187,000 to $152,000. This will directly impact the acquisition of talented software and computationally focused researchers that are needed for life sciences at high profile centers. These centers need to try to lure experts away from the large hyperscalers and cloud or social media companies who have significantly more liberal compensation packages. It’s all extremely complicated. However, it is important to remember in all discussions about life sciences funding, that while the initial human genome consumed $1 billion in federal investment, it gave a $140 billion return to the private sector, and that’s only so far. That’s significant leverage, the potential knock on effect of advanced computing in that financial equation should also not be dismissed lightly.
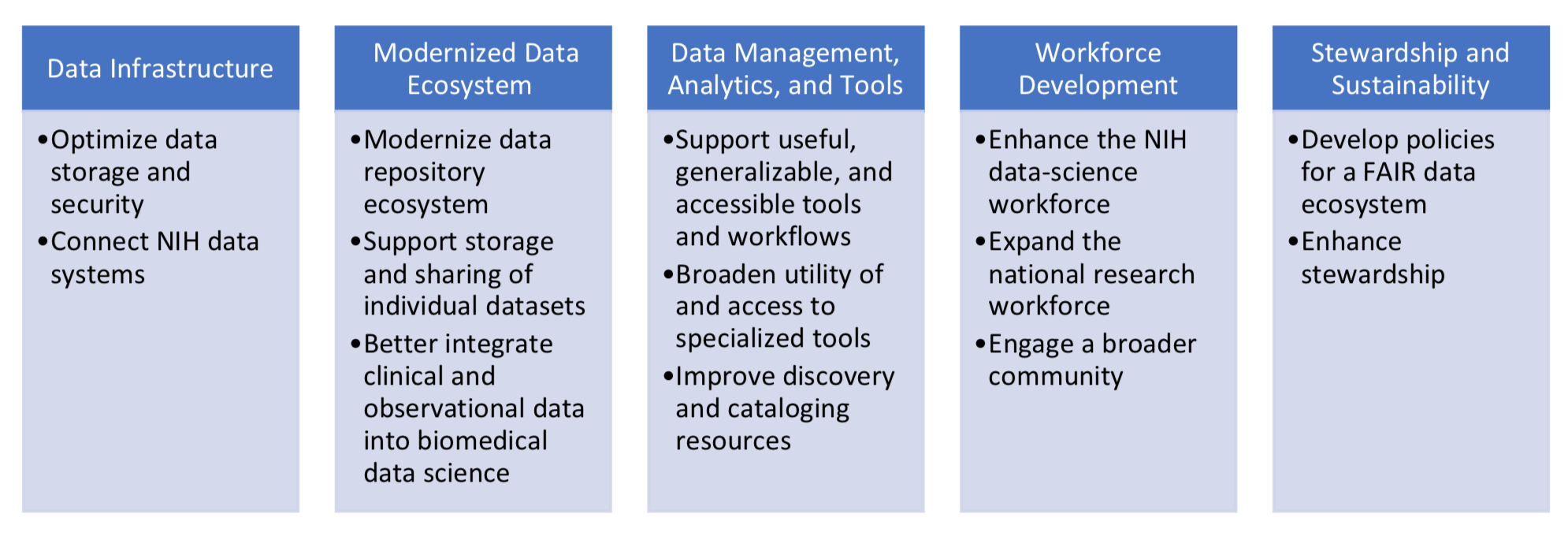
The Five Step Data Science Plan
So what exactly under these critical constraints are the NIH planning to do to support data science then? They essentially present a five step path to support computing and storage and systems. The five are: data infrastructure, a modernized data ecosystem, data management and analytics and tools, workforce development and finally stewardship and sustainability. Two stand out pieces are clearly around data infrastructure and the workforce development. These also align directly with the vision coming out from the Broad Institute. Better software and better tools with the ability to expand and enhance the people needed to build, write and understand them. It is a solid plan.
The 31 page report also directly confronts the issue of exascale computing in life science by clearly saying, “At the exascale level of computing speed, supercomputers will be able to more realistically mimic the speed of life operating inside the human body, enabling promising new avenues of pursuit for biomedical research that involves clinical data.”
“Mimic the speed of life operating in the human body.” Let that just sink in there for a moment. The report continues to then highlight specific use cases that will be both early adopters and the drivers of exascale computing, “These data-intensive programs may well be among the earliest adopters and drivers of exascale computing: They include the All of Us Research Program and the Cancer Moonshot components of the Precision Medicine Initiative, the Human Connectome project, the Brain Research through Advancing Innovative Neurotechnologies (BRAIN) initiative, and many others.”
Philippakis in his opening anthem at the Broad specifically called out the All of Us research program, and showed yet another one of those terrifying hockey stick graphs. There is good reason to be concerned about exactly how much computer time will be needed across such programs. There is a clue right there in the name, “All of Us”. The project quite literally aims to “collect data from one million or more people living in the United States to accelerate research and improve health. By taking into account individual differences in lifestyle, environment, and biology, researchers will uncover paths toward delivering precision medicine.” That’s a whole lot of data. Basically a million human genomes, plus all the rest of it for the lifestyle analysis, plus this is all but one single project amongst thousands that are all taking place at the same time under the auspices and guidance of the NIH. Every single one of them will need some level of computing. Some will require such massive amounts of computing that whole new methods will be need to be developed. All of this needs to happen under strict budget controls.
Computing And Storing
Houston, we see the problem. This is exactly why we cover the new machines and advanced processing capabilities that are going to be needed to solve these challenges at a massive scale and also not need thousands of new power stations to be built in an attempt to run them. The problem is effectively two-fold, nowhere near enough computing or enough skilled humans are available to be able to program them, this is highlighted in the overarching strategy for modern life sciences research.
We first looked at the application performance of future of exascale shaped machines like “Summit” at Oak Ridge National Laboratory in March 2018. Recently Summit was finally released with a great deal of fanfare and statements about exaops performance being delivered by the new 207 petaflops machine. The comment has caused a little stir in the community about whether this was true to the pure design goals of “exascale,” with many meaning exaflop rather than exaops performance metrics. However, the Oak Ridge National Laboratory leadership computing facility recently showed the impact of the Summit machine on a specific life sciences algorithm, CoMet. By effectively turning down the precision carried by the algorithm as it executed and then leveraging lower accuracy, but faster tensor cores, they were able to see significant speed up and also manage to execute the same workload to deliver accurate science. These are encouraging results. Dan Jacobson, an ORNL computational biologist said, “the approaches that we take with CoMet allow us to discover fundamental biology.” It is a valid point, life science algorithms have always had an interesting shape and optimal placement of the right algorithm at the right time on the right facility is how real progress will be made.
But what about the data?
Philip Bourne famously explained in a 2016 vision document for BD2K that total data from NIH projects is estimated at 650 PB and that in 2015, only 12 percent of the data described in published papers lives inside of recognized archives. Meaning that 88 percent of all life science data is considered dark. Read it again: 88 percent of data is dark, it can’t be found. Between 2007 and 2014, the NIH also spent $1.2 billion on extramural activities to support data archives. Bourne was the first associate director for data science at the NIH before moving to be the Stephenson chair of data science and director of the data science institute at the University of Virginia.
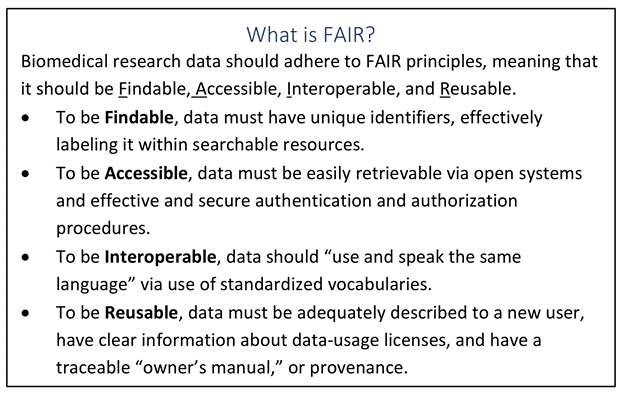
The main part of the NIH strategy is also underway as they are searching for a new chief data strategist and director for the office of data science strategy. Fundamentally, this has to be one of the most challenging positions today in the life sciences, applications will be reviewed starting on the 9th of July, 2018, so watch this space. However, what is going to be crystal clear and critical is that the FAIR principles will be fully baked into this new role and that of the entire organization. FAIR is now such a commonly used vocabulary to help make sure that data is findable, accessible, interoperable and reusable. No more dark data, 88 percent that Bourne stated cannot be the new normal. From a distance, it is almost as if public cloud computing with their vast shared, highly indexable and sharded high performance data stores and FAIR were almost designed to fit together like the proverbial bricks in the Lego system. Each piece does fit, it can be reused and there is some significant intention here in terms of setting the data strategy for the NIH.
A Cloudy Future
Because of this challenge with FAIR and data, the NIH also clearly needs to have a strong cloud game, especially for their plans to more fully integrate storage and regain control of their storage. The NIH report clearly states, “Large-scale cloud-computing platforms are shared environments for data storage, access, and computing. They rely on using distributed data-storage resources for accessibility and economy of scale – similar conceptually to storage and distribution of utilities like electricity and water.”
Clearly the NIH has a solid grasp on the core technology, and they then continue with, “Cloud environments thus have the potential to streamline NIH data use by allowing rapid and seamless access, as well as to improve efficiencies by minimizing infrastructure and maintenance costs.” This is an important line on the overall economies of scale and potential, this is good. To seal the deal the group continues with, “NIH will leverage what is available in the private sector, either through strategic partnerships or procurement, to create a workable Platform as a Service (PaaS) environment.” Well then. So that’s all pretty clear in terms of the overall approach, there will certainly be a whole lot of cloud in the future of life sciences.
So there we have it. There will be way more data, a whole lot more of it, massive amounts of computing will be needed to do anything with it, and it looks like the majority of it will sit on some sort of cloud infrastructure. We also still actually don’t have enough skilled humans to build the software to run it, but everyone seems to be super focused on working on that particular issue in parallel.
This is a good time for life sciences and research computing in general, we agree with Philippakis, the next decade of advanced HPC and life sciences really will be exciting. And yes, you just ain’t seen nothing yet.



Be the first to comment