
When it comes to DNA analysis, large-scale pattern matching algorithms are constantly being refined to add a performance and efficiency boost to speed the time to result and accordingly, the practical implementation of fast analysis for more accurate, timely, personalized medical care and research.
Such pattern recognition algorithms are central to understanding the key differences and evolutionary influences behind samples, but require very large comparative efforts—a task that is both data and compute-intensive. Although CPU-only systems have a very strong foothold in the DNA sequencing and analysis field, over the years there has been a wealth of research around the viability of using both high-end CPUs coupled with accelerators to speed the DNA sequencing pipeline.
For instance, there have been many studies offering benchmarks and performance assessments for refining DNA sequencing models to fit the CUDA platform for an NVIDIA GPU boost with success. But oddly enough, even though the Intel Xeon Phi coprocessor has risen through the supercomputing ranks to be a comparable accelerator option for demanding workloads, there is relatively little about its use for genome analysis, despite the fact that there are algorithms that are already well-suited for it on the pattern matching front.
On that note, pattern matching has historically been considered a solid fit for GPU acceleration, with some algorithms getting well over 30X to over 100X the performance of their sequential code kin. As described here in the context of pattern matching for deep packet inspection at a 30X boost, or here for actual DNA sequence pattern matching for nearly 100X, pattern matching has many use cases that map well to the GPU. So why is there comparatively little in the way on this for the Xeon Phi, which has been designed with architectural features to make it a perfect fit as well?
This is one of the questions a Swedish team set about to answer in their evaluations of Xeon Phi over standard CPU for genetic pattern matching. Among such codes for genetic pattern matching is a finite automata-based parallel algorithm, which was modified to exploit both the SIMD and thread-level parallelism inherent to the Xeon Phi. Following work on the algorithm, they were able to show up to a 10X improvement over the sequential implementation on a Xeon E5-series processor for DNA pattern matching across tests using human and other animal DNA.
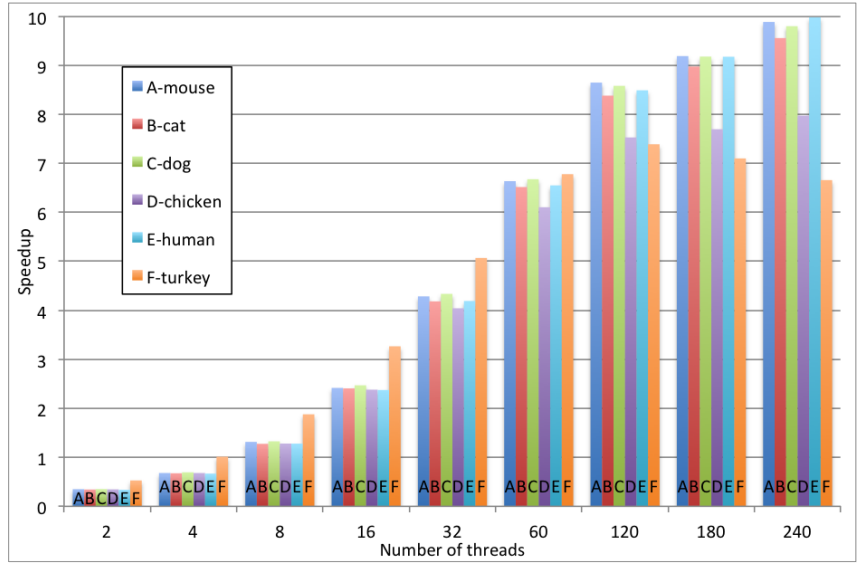
“With respect to the SIMD parallelism, the algorithm implementation uses the potential of 512-bit vector registers of the Intel Xeon Phi architecture for the transition function of the algorithm,” they note. Furthermore, there are several optimizations within the algorithm to boost performance that are detailed in the full paper. Interestingly, the best scalability the team observed among the different DNA types used for the benchmark (which included chicken, turkey, dog, cat, and others) was with the human sequence, which was the largest used in the experiment.
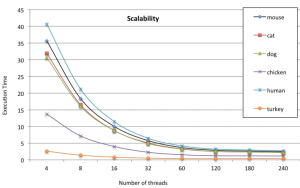
This is where the 10X speedup was observed using 240 threads (compared to a sequential version running on a Xeon E5-series processor. As one might imagine, the next generation Knights Landing processor—the extension to the Xeon Phi set to come out next year—will provide an even greater potential boost, which the researchers say is their next target for future benchmarks.
The real question this research generates is why there are not more research results for the Xeon Phi for genomics analysis and related pattern matching algorithms. One can assume that with the launch of the Knights Landing chips a fresh wave of benchmarks and use cases will occur, but there is no comparison between the volume of GPU to Xeon Phi research in genomics in particular.
As The Next Platform reported earlier in the summer, the Knights Landing chip is expected to deliver at least 3 teraflops of double-precision floating point performance and twice that at single performance – that predictability will be a big asset, because GPU accelerators often are better at DP or SP, depending on the make and model. It is not clear what clock speeds or how many cores will be active to attain this performance level with Knights Landing, and it is quite possible Intel itself is not sure yet. It will also have up to 16 GB of high bandwidth memory (HBM) on the package (the near memory) and another 384 GB of DDR4 memory (this is the far memory) for data and applications to frolic in. The DDR4 memory will deliver 90 GB/sec of bandwidth into and out of the Knights Landing’s cores, which have two vector math units each, and the near HBM memory will deliver more than 400 GB/sec of memory bandwidth.
This will no doubt lead to a much bigger leap in application performance than the move from the current 61-core Xeon Phi cards, which were rated at a little more than 1 teraflops at double precision, to the Knights Landings, which have triple that raw floating point performance. With the processor and co-processor together at last, the use cases and research that did not emerge strongly for the first generation Phi could be overwhelming once in production.




maybe its simply because GPUs are faster & cheaper and meanwhile they are easier to program when looking for serious utilization since the CUDA/OpenCL frameworks expose the parallell hardware and vectorization units efficiently…. futhermore, this generation of GPUs already provide up to 8 TFLOP/s of processing power (more than knights landing will have), meanwhile soon to be next gen. GPUs expect to double that performance… there simply arent any strong arguments for using a cpu in this types of workload.
Well that might be because Intel’s support for the 1st gen XeonPhi was dyer at the beginning unless you were a top tier HPC shop. It has improved in the mean time and one can hope it will improve even further with KL
There are two different types of genomic sequencing;
1. Matching to a reference genome
2. De novo sequencing
The first matches a bunch of DNA fragments to locations on the pre-existing “reference human genome”. This takes about an hour on a PC. It can be sped up by GPUs. It is the technique used to do the $1,000 genome sequences.
The second assembles about 10- 30 million DNA fragments into a complete genome. It is similar to assembling a giant jigsaw puzzle. Each segment can be from 200-1000 base pairs long. Every fragment must be compared to every other one. The solution is usually framed as a Big Data graph analytics problem and can takes weeks to resolve.
Due to the massive memory requirements GPUs are generally not helpful.
De novo sequencing is what is required to analyze cancer tumors since the tumor DNA contains so many insertions, deletions, and mutations that comparing to the reference genome is not helpful.
Billions are being spent on this problem because its solution leads to the ultimate cure for cancer.
this gen GPUs have up to 12 GB of RAM while 2016 GPUs will have up to 32GB (HBM) or more for HPC, and they can also share nenory with the host system directly.
Animal and human genomes are relatively small when compared to plant genomes like wheat, corn. We had a customer asking for 20TB of memory to do their genome job. Only specialized numa machines are up to such tasks today …